Product Quantization for Vector Similarity Search (+ Python)
Chapters
0:0 Introduction2:42 Dimensionality Reduction
4:50 Product Quantization
9:9 Python Code
18:31 Coding
21:16 Visualizing
22:26 Quantizing
Transcript
I will come to the next video in our series on similarity search. We're going to be covering product quantization, which is a very effective method for reducing the memory usage of our vectors. Now, I'll just show you this very quickly. So we, in the first column over here, we have the recall speed and memory for a dataset of size 1 million.
So we're using the SIFT 1M dataset here. And the dimensionality of the vectors in there is 120. It's a very small compared to a lot of vector sets out there. Now, the memory usage of that, when we just saw them as they are, is a quarter of a gigabyte, just over, which is pretty big.
And you think this is a small dataset, small dimensionality, so this is already pretty significant. And when you start getting into high dimensionality or just larger datasets, it quickly becomes very unmanageable. So product quantization, or PQ, allows us to reduce that significantly. So in this example, so the tests that we run here, we reduce the memory usage by 97%, which is pretty significant.
So from a quarter of a gigabyte to 6.5 megabytes, it's like nothing. And then the speed increase is pretty significant as well. So just under a six-fold speed increase there. So pretty good. Obviously, the recall also decreases a fair bit. So that's just something that we need to be careful with.
But at the same time, there's different parameters that we can use in order to fine-tune that. So what we're just going to cover in this video is the logic behind product quantization. There will also be another video, which will be following this one, where we'll have a look at product quantization in FICE.
And we'll also have a look at a composite index of product quantization and inverted files, so IVF. And with that, we can actually make the speed even faster. The memory goes up a little bit to, I think it's like nine megabytes, so pretty minimal. But the speed increase is very significant.
It's like a 92 times speed increase over a flat index. So it's super fast. But we'll be covering that in the next video. For this one, I'm just going to work through what product quantization is and how it works. So we'll start with the example of dimensionality reduction, which is what you can see here.
So we have this S value up here, which is kind of like the scope of possible vectors that we can have, so all along here. In the middle here, I've highlighted a single vector. So this would be X, our vector. And D over here is, of course, our dimensionality of that vector.
So typically, well, in this CIF-1M data set, for example, we'd use a dimensionality of 128. Dimensionality reduction is something like PCA, for example, where we would just reduce the dimensionality there. And we would try to maintain the geometric or spatial properties of that vector. Even though it has less dimensions, we would still try and maintain that.
So vectors that were close to each other before with a high dimensionality should be close to each other again in low dimensionality. Now, what we have here is not dimensionality reduction. This is quantization. Now, quantization is a very generic term. And it focuses on any method where we're trying to reduce the scope of our vectors.
Now, in this case, so beforehand, the scope is typically either a very large number or technically infinite. So if we're using floats, for example, the number of possible vectors that you can have within that space is usually pretty big. You may as well say it's infinite. Now, in this case, what we do is reduce that.
So we reduce the possible scope into a more finite set. Then we may be going from something like a practically infinite scope to something like 256 possible vectors. That would be a very small number, but it's just an example. Then notice that the dimensionality does not necessarily change. It can change with quantization, but that's not the point of quantization.
The point is to reduce the scope of our possible vectors. Now, at a very high level, let's just kind of cover what product quantization is doing. So what we have here, this big blue block at the top here, this is our initial vector x. Now, we'll say dimensionality, this is 128.
So D equals 128. Now, the number of bits required to sort this vector, let's say it's every value is a 32-bit float. So we have 32 bits multiplied by the dimensionality is 128. So in total, that gives us 4,096 bits that we need to sort a single vector. Now, that is pretty significant.
So what we are going to do with PQ is try our best to reduce that. First thing we do with product quantization is split our vector x into subvectors, each one represented by U. So U to a subscript of J, where J is a value from 0 up to 7 in this case, representing the position of that subvector.
Now, the next step is the quantization of product quantization. So the next step, we process each one of these subvectors through a clustering algorithm. Each of these clustering algorithms is specific to each subspace. So it's specific to J equals 0, 1, 2, 3, or so on. And within each one of those clustering sets, we have a set of centroids.
Now, the number of centroids, we set that beforehand. So when we're building our index, we set the number of centroids that we want. And the more centroids we have, the more memory we use, but also the more accurate the index becomes. Because you think you have, say you have two centroids in one vector subspace.
Your subvector will have to be assigned to one of those two centroids. And in the case that we only have two centroids, neither of them could be particularly close to our vector or subvector. So that average distance between subvectors and the clusters that they're assigned to is called the quantization error.
And for good accuracy, good recall, good results, we want to minimize that quantization error as much as possible. And we do that by adding more centroids. So if we compare our cluster of two centroids to a set of clusters where we have, let's say, 200 centroids, the chances are we're going to find a centroid that is more closely fitting to our subvector in the 200 centroid algorithm than the one with just two centroids.
So that's the logic behind it, sort of the memory usage and accuracy trade-off there. Obviously, the more centroids we have, the more data we store in our index to cover the full scope of all those centroids. Now, once we've assigned a centroid to our subvector, we map that centroid to its unique ID.
So every centroid within this index will have a unique ID. That unique ID is a 8-bit integer, typically. So we've now gone from-- so originally, we had a vector x of dimensionality 128. And each value was a 32-bit float. So the number of bits we use there, 4,096. Now we only have a ID vector, which contains eight 8-bit integers.
So that's 8 multiplied by 8. In this case, we have 64 bits. So we've compressed our 4,096-bit vector into a 64-bit vector. And that's what gets stored in our index. Now, there's also something called a codebook, which maps each one of those IDs back to the reconstruction or reproduction values, which are those centroid vectors.
But of course, because we've minimized the scope there, we don't need to store that many of those mappings. So say our scope is 256, we only need to store 256 of those mappings. So this is how product quantization can be so effective. Now, we'll go through some more visuals.
But I want to actually write this out in code as we go along. For me, at least, writing out in code makes it much easier to at least logically understand the process. So there are a few variables that we need to define here. So we have this vector up here.
It's very small. It's using integer values. The data types here don't matter. We just want to build out the process of product quantization. This is not an effective implementation or anything like that. We just want to understand the process and logic behind it. So the first thing is, how many subvectors are we going to convert our x full vector into?
In this case, I want to create four subvectors. And we also need to get the dimensionality of x as well, which is just the length of x. Now, we have those two. And the first thing we need to do is make sure that d is divisible by m, because we need equally sized subvectors.
So what we do is rewrite assert that d is, in fact, divisible by m. So we just write this. And if that is the case, we don't get an error. So if I just add a 1 in there, I'm going to throw it off. We get this assertion error.
So we just add that in there to say, OK, d is definitely divisible by m. So that's good. We can continue. And what we want to do is say, OK, what is the length of each subvector? It's going to be equal to d divided by m. And we'll have to convert that over to an int as well.
So we'll just do that. And let's see what we get. So each subvector will be of size 3. Now, let's build that. So our subvector, or our set of subvectors, will be assigned to u. And what we're going to do is go x from row to row plus d, d underscore, for row in range 0 to d.
And we're going to take it in steps of d, d underscore. OK. And then let's see what we get. And see that we now have our subvectors. Now, what does that look like? Let's visualize that. So here we have that vector. So d up here is our 12. And we're splitting that vector x of dimension 12 into four subvectors.
So m equals 4 down here. We'll create four subvectors. Now, d over m, which is what we calculated before, produces this d star. Now, in the code, we can't write the star. So I've changed it to d underscore. And the value of that is 3. So each of our subvectors has a dimensionality of 3.
So here we have that d star dimensionality of 3 here. These are our subvectors. We have 4 in total. So that is equal to m. Now, next thing I want to do is answer k. So k is the number of possible values that we're going to have in our data set.
So we are going to do 2 to the power of 5. And we print it out. And we get 32. So k is going to be equal to 32. Now, the one thing we need to make sure of here is this k value, so the number of possible centroids, that is going to be shared across our entire set of subspaces or subvectors.
So we need to make sure that that is divisible by m again. So we do the same thing again. We say assert that k is, in fact, divisible by m. If it is, we can go on to get k star or k underscore, which is going to be k divided by m.
OK. And let's have a look at what that is. So this means that with that, we should also make sure that's an integer, like so. OK. So what that means is that we are going to have 8 centroids per subspace. So each one of these subvectors here will have the possibility of being assigned to one of the nearest 8 centroids within its subspace.
And in total, across all of our subvectors, of course, we have a total of 32 possible centroids. Now, what that 32 there means is that we will have a codebook. So a codebook is simply a mapping of our centroid vectors. Or, in fact, let's do the other way around.
It's a mapping of centroid vector IDs. So each one of the centroids that we haven't created yet but we will create will be assigned a unique ID. And it maps us from the unique ID to the actual centroid subvector. So that's where later on we call those centroids reproduction values.
Because later on, when we're searching through our index, our index is storing those 8-bit integers. But obviously, we can't compare those 8-bit integers to an actual vector that we're searching with, our query vector. So what we do is we map each one of those 8-bit integers, which are the IDs, back to their original centroid values.
Not to the original subvector values, but to the original centroid values. And that's what our codebook is for. So this k value, this 32, means that we will have a total of 32 mappings in our codebook. Now, I know that can maybe seem confusing. So let's have a look at what that actually looks like.
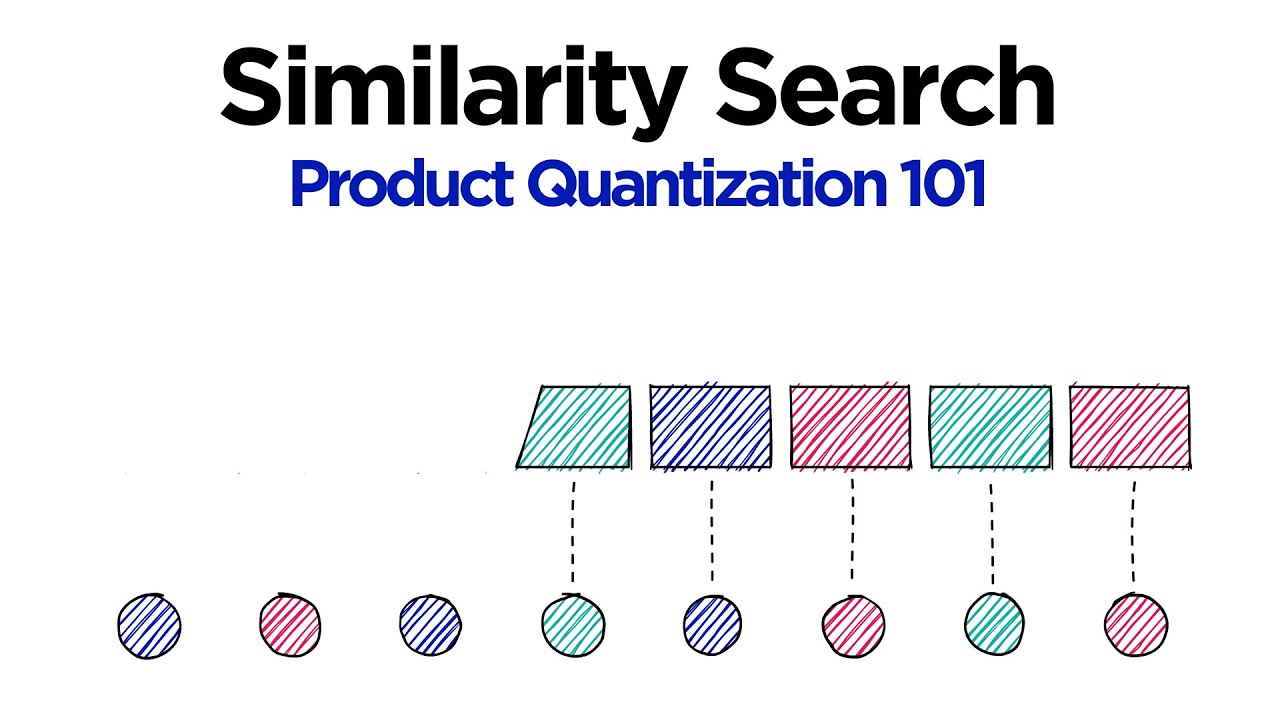
So over here, we have u and our subvectors. Here, we would have, let's say, j equals 0, j equals 1, 2, 3. And over here, what we have is our reproduction value. So those centroid or those cluster centroids. Now, in this case, we have these three centroids. In reality, we set k equal to 8.
So we should actually see not 3 centroids here, but we would actually see 8. So we have 1, 2, 3, 4, 5, 6, 7, 8. So in reality, there would actually be that many centroids in there. And each one of those centroids can be referred to as c, j.
So in this case, j would be 0 and i. So we would have, let's say, this centroid is i equals 0, this centroid is i equals 1, this centroid is i equals 2, and so on, all the way up to, in this case, 7. So 0 is 7, giving us the 8 centroids there.
So to get to this centroid here in our codebook, so this c represents our codebook, we would go c, 0, 0. Or this one over here, this number 2, would be c, 0, 2. Let's say down here, this green centroid, let's say that is i equals 4. So that means that to get to that in our codebook, we would say c, 3, 4.
Okay, so that's just the notation behind it. So our quantized subvector is one of these centroid subvectors. And each one of our reproduction values, or those subvectors, the quantized subvectors, will be assigned a reproduction value id, which is simply what we have here. So this c, j, i. So in this case, this value here may be, it may be the value 0, 8 integer 0.
And if we started counting through all of our codebook, that would relate to c, 0, 0. Now if we were to go, let's say down here, this is number 10, the integer value of 10. Now that if we count through, so j is going to go up to 7, and that's our integer value of 7.
And it's going to get reset because we're going to take j from 0 to 1, which is going to reset our i counter. So it will go from c, 0, 7. And the next one in our codebook will be down here, which will be c, 1, 0. So this actual integer value in our id vector, that would be represented by the value 8.
So this value here would be 8, so we add one more. We go to 9, so that would be c, 1, 1. And 10 would be c, 1, 2. So c, 1, 2. Okay, and that's how we represent those. And we refer back to those original centroid subvectors within the reproduction values codebook.
But for now, we don't have those centroids, so we need to create some. So what I'm going to do is from random, import random int. C is going to be our overall codebook or list of reproduction values. And we're going to say for j in range m, so we're looping through each of our subspaces here, we are going to initialize cj.
Okay, and then in here, we need to store eight different centroid vectors or reproduction values. And of course, we refer to those as i, so position those as i, for i in range. And here, we are looping through k_. So in our case, that would be equal to 8.
So we run through that 8 times. Here we have m, so you think 4. We loop through 4 j's, and we loop through 8 i's for each one of those. So in total, we get that 32 value, which is our k value. And what we want to do is say we're going to say cji is going to be equal to, we want to say randint from 0 up to, it's going to be our vector space, our cluster vector space.
So we're just going to set it to the maximum value that we have in here. So we have a 9. So we set that. So these are the centroid values, remember, not the ids for just underlying in range. Here we have our dimensionality of each subvector. And what we want to do is just append that to cj, cji.
And then outside that loop, we want to append cj to c. Okay, so we've now just created our clusters, which we can see in here. So each subspace, so each j has a total of 8 possible centroids. So we have 0 or 1, 2, 8 here in each one.
And we don't forget we have m of those subspaces, so we should see 4 of them, although it does put out here, but this is in fact just the fourth one here. Now I think it's pretty cool to be able to visualize that. So I'm just going to copy and paste this code across.
You can see here, you can copy if you want. It will be also in the notebooks that will be linked in the description of the video. And we're just going to visualize each of our centroids within each of those subspaces. So this is c, where j is equal to 0, j is equal to 1, 2, and 3.
And these here are our centroids. Now typically, of course, we would train these, but we are not going to train them. We just want to create a very simple example here. So we're not going to go that far into it. Okay, so I mean that's everything we see there.
And now what we need to do, so we've produced those reproduction values, all those centroids, and now what we need to do is actually take our subvector and assign it to the nearest reproduction value or centroid. Now how do we do that? We simply calculate the distance between our subvector and each of its equivalent reproduction values or centroids.
I'm sure it's going to get annoying if I keep saying reproduction values or centroids, so I'm just going to call them centroids from now on. It's so much easier. So we are going to calculate, we're going to identify the nearest of our centroids to our specific subvector. So I'm just going to copy these functions in.
The top here, we're just calculating the Euclidean distance. It's pretty straightforward. If you need to just Google Euclidean distance, the form is pretty straightforward. And then here we're just using that Euclidean distance and looping through each of our values, so the K_, so each of the centroids within our specific J subspace.
So all we need to do there is pass a specific J subspace and our specific subvector to that function. And to do that, all we want to do, I'll just also note that what we're doing here, these are the index positions, so the high positions, and we're looping through and saying if the new distance that we calculated between, you see here we're accessing that specific centroid, if that is less than the previous one between our subvector and that centroid, then we assign the nearest ID to that vector, not necessarily distance.
We don't care about distance, we just want to see the nearest ID to nearest IDX, which is what we return. So what we're going to return there are in fact the I values for those centroids. So first thing we're going to do is initialize our ID's vector, so here we're actually going to build that 8-bit integer IDs.
In this case, there's not going to be eight of the values, there will be four, because the length of this vector is always going to be equal to M. So we have 4J in range M, I is equal to the nearest between CJ and UJ. I want to say ID's append I.
And then let's see what we have. Okay, so that is our quantized subvector, so each one of these IDs here represent one of our centroid positions. So if we wanted to get those centroid positions, so in this case I'm going to call them the reproduction or reconstruction values, we would do this.
So this is going to be our reconstructed vector. I'm going to go between, so we're going to go for each J in range M, again remember, we're going to say CJI, so the reconstruction value, is equal to C for J. And then the I position is whichever value we have in here, so we need to say ID's J.
Okay, and then we're going to do Q, extend, CJI, and now let's have a look at what we get for Q. So this is our fully reconstructed vector. Now earlier on I said that we get the quantization error, and we typically measure that using the mean squared error, so this function here, you can see it there.
So what we can do is we can measure the error, or mean squared error, between our quantized vector and the original vector. So all we do here is we go mean squared error, X, and Q. And here we get the total squared error, mean squared error, between our original vector and the quantized version.
Through sort of increasing the M value, or increasing K, we should be able to minimize this, and therefore improve the performance of our index. So I have one final thing to show you, which is how the search works in PQ. So on the right we have just a single ID vector.
We would use our codebook, C, so that they would each go into this, like so. And from there we would output our quantized subvectors. So these are their reproduction values. Now on the left we have our query vector, and we would split that, of course, like we did before with our initial vectors.
So we would split that, and we would get this. Now, what we would do is we calculate the Euclidean distance between these, and what we would get is we would simply add all of these up. So we'd take the product of all of them, hence why this is called product quantization, and the total distance would be all of those added together.
Okay, let's say this. Now we would do that for all of our vectors. So this is a single, so this is like QU. That's a single quantized vector, or set of quantized subvectors. We need to do that for all of them. And then we just find one that produces the lowest or the lowest or the smallest product distance.
Now, as well, it's worth noting, because we're taking the product, this isn't really the distance between the two vectors, between U or the quantized version of U and XQ, but it's almost like a proxy value for that. And then that's what we use to find, to identify the nearest vector.
Now, I'm not going to go into the code behind this because it's quite long, but I am going to, I'm definitely going to record that and just leave it as like an extra or almost bonus video if you do want to go through that. But it is, it's pretty long, so I wouldn't say you necessarily need to.
But in the next video, anyway, we're going to have a look at how we implement all this in FICE, which is obviously much more efficient. And it is in that video that we will also introduce the IVF and PQ CompSetIndex, which just allows us, if you have a look at the speed here, to really reduce the speed an insane amount.
It's like a 90 times increase in the speed of our search. So it's pretty cool. But yeah, that's it for this video. So thank you very much for watching, and I will see you in the next one.